Manual install (install from source code)
-
Install ISEScan
-
Download the latest ISEScan from https://github.com/xiezhq/ISEScan/releases, e.g. Source code (tar.gz).
-
Uncompress the .zip (or .tar.gz) file.
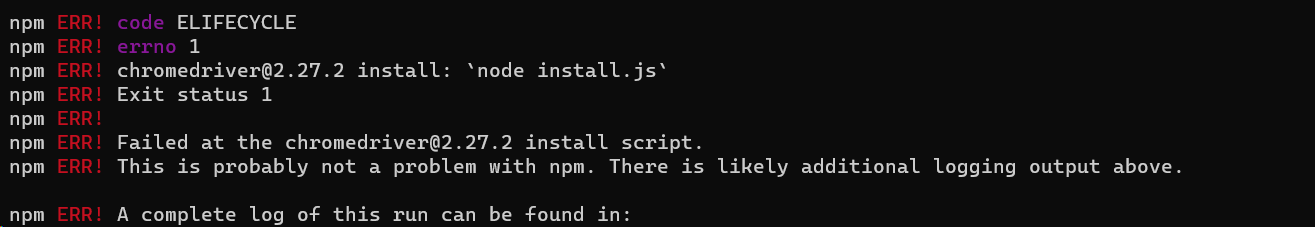
unzip v1.7.2.2.ziptar -zvxf v1.7.2.2.tar.gzThis will create a ISEScan folder, e.g. ISEScan-1.7.2.2. You need to go to ISEScan folder to configure and run it.
cd ISEScan-1.7.2.2
-
-
Install dependencies before you run ISEScan
- Python 3.3.3 or later 可以使用conda安装 挺快
- numpy-1.8.0 or later 这一步不用安装 因为在安装fastcluster时候会自动安装
- scipy-0.13.1 or later 使用pipin stall -i https://pypi.douban.com/simple/安装
- fastcluster, latest version recommended, fastcluster · PyPI
- FragGeneScan1.30 or earlier, (The .faa file output by version1.31 is not compatible with ISEScan!), FragGeneScan
- HMMER-3.1b2 or later, HMMER3.1使用 – 简书 HMMER conda安装hen’s HMMER
- BLAST 2.2.31 or later
- SSW Library, the latest version is not tested with ISEScan and the tested version of SSW library is shipped with ISEScan, please find it at ssw201507 subdirectory.
- To use the shipped SSW library in ISEScan, please go to ssw201507 and then compile the codes by gcc:
cd ssw201507 gcc -Wall -O3 -pipe -fPIC -shared -rdynamic -o libssw.so ssw.c ssw.hcp libssw.so ../ export LD_LIBRARY_PATH=/home/xiezhq/projects/ISEScan-1.7.2.2:$LD_LIBRARY_PATHIn command
export LD_LIBRARY_PATH=/home/xiezhq/projects/ISEScan-1.7.2.2:$LD_LIBRARY_PATH, please replace/home/xiezhq/projects/ISEScan-1.7.2.2with the actual path of libssw.so on your computer! - biopython 1.62 or later (required by SSW library)
-
Add the required packages to your $PATH before you run ISEScan
- Add to $PATH the paths pointing to run_FragGeneScan.pl, phmmer, hmmsearch, blastn, blastp, makeblastdb
export PATH=$PATH:/apps/inst/FragGeneScan1.30:/apps/inst/hmmer-3.3/bin:/apps/inst/ncbi-blast-2.10.0+/binIn command export above, please replace
/apps/inst/FragGeneScan1.30,/apps/inst/hmmer-3.3/binand/apps/inst/ncbi-blast-2.10.0+/binwith the actual paths of FragGeneScan, HMMER and BLAST on your computer!
kobas
conda create -n kobas python=2.7
#使用activate激活环境直接安装
conda activate kobas
conda install -c bioconda kobas==3.0.3metagenemark
MetaGeneMark-2-exp/reproduce.pdf at master · gatech-genemark/MetaGeneMark-2-exp · GitHub
$ conda install -c bioconda metaphlan
mgrast
motusGitHub – HRGV/phyloFlash: phyloFlash – A pipeline to rapidly reconstruct the SSU rRNAs and explore phylogenetic composition of an illumina (meta)genomic dataset.
nt nr
NR/NT,Taxonomy和RefSeq——三种NCBI常见数据库 – 大漠胡天 – 博客园 (cnblogs.com)
cd ${db}
mkdir -p NCBI_nt && cd NCBI_nt
wget -c “ftp://ftp.ncbi.nlm.nih.gov/blast/db/nt.*.tar.gz”
mkdir -p NCBI_nr && cd NCBI_nr
phyloflash 从头安装
YAMDA/README.md at master · daquang/YAMDA · GitHub
conda install -c bioconda pybedtools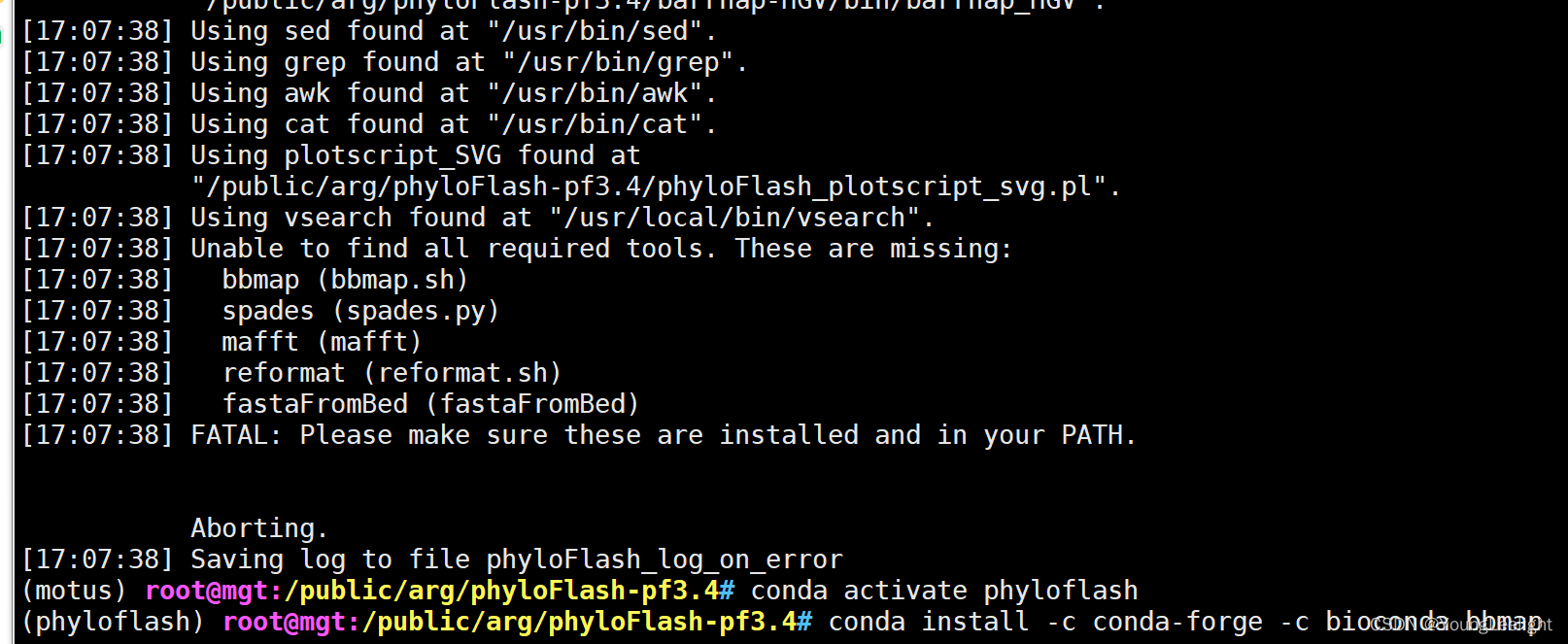
https://github.com/daquang/YAMDA/blob/master/README.md
bioinformatics-cafe/fastaFromBed at master · dariober/bioinformatics-cafe · GitHub
3、添入环境变量
echo ‘export PATH=~/.aspera/connect/bin/:$PATH’ >> ~/.bashrc
source ~/.bashrc
plasflow 自建频道 自建channel
https://github.com/smaegol/PlasFlow
qiime2
QIIME 2 command-line interface (q2cli) — QIIME 2 2022.11.1 documentation
Installing QIIME 2 — QIIME 2 2022.11.1 documentation
signalp5 signalp6
GitHub – chenxi-zhang-art/signalP
tar –xzvf signalp-5.0b.Linux.tar.gz
https://github.com/fteufel/signalp-6.0/blob/main/installation_instructions.md
Description
A placeholder package for SignalP v5.
Due to license restrictions, this recipe cannot distribute signalp5 directly. Please download signalp-5.0b.Linux.tar.gz from: https://services.healthtech.dtu.dk/services/SignalP-5.0b/9-Downloads.php#
and run the following command to complete the installation:
$ signalp5-register signalp-5.0b.Linux.tar.gz
This will copy signalp5 into your conda environment.pip install signalp-6-package/ -i https://pypi.douban.com/
simple/
plasmidfinder
https://github.com/kcri-tz/plasmidfinder
魔鬼报错
conda install -c bioconda -c conda-for
ge plasmidfinderCollecting package metadata (current_repodata.json): done
Solving environment: done
## Package Plan ##
environment location: /share/app/miniconda3/envs/plasmidfinder
added / updated specs:
- plasmidfinder
The following packages will be downloaded:
package | build
---------------------------|-----------------
biopython-1.80 | py311hd4cff14_0 2.9 MB conda-forge
boost-1.80.0 | py311h59ea3da_4 358 KB conda-forge
boost-cpp-1.80.0 | h75c5d50_0 17.4 MB conda-forge
cgecore-1.5.6 | pyh3252c3a_0 34 KB bioconda
kma-1.4.9 | h7132678_0 261 KB bioconda
numpy-1.24.2 | py311h8e6699e_0 7.4 MB conda-forge
plasmidfinder-2.1.6 | py310hdfd78af_1 1.7 MB bioconda
tabulate-0.9.0 | pyhd8ed1ab_1 35 KB conda-forge
------------------------------------------------------------
Total: 30.1 MB
The following NEW packages will be INSTALLED:
_libgcc_mutex conda-forge/linux-64::_libgcc_mutex-0.1-conda_forge
_openmp_mutex conda-forge/linux-64::_openmp_mutex-4.5-2_gnu
biopython conda-forge/linux-64::biopython-1.80-py311hd4cff14_0
blast bioconda/linux-64::blast-2.5.0-hc0b0e79_3
boost conda-forge/linux-64::boost-1.80.0-py311h59ea3da_4
boost-cpp conda-forge/linux-64::boost-cpp-1.80.0-h75c5d50_0
bzip2 conda-forge/linux-64::bzip2-1.0.8-h7f98852_4
ca-certificates conda-forge/linux-64::ca-certificates-2022.12.7-ha878542_0
cgecore bioconda/noarch::cgecore-1.5.6-pyh3252c3a_0
icu conda-forge/linux-64::icu-70.1-h27087fc_0
kma bioconda/linux-64::kma-1.4.9-h7132678_0
ld_impl_linux-64 conda-forge/linux-64::ld_impl_linux-64-2.40-h41732ed_0
libblas conda-forge/linux-64::libblas-3.9.0-16_linux64_openblas
libcblas conda-forge/linux-64::libcblas-3.9.0-16_linux64_openblas
libffi conda-forge/linux-64::libffi-3.4.2-h7f98852_5
libgcc-ng conda-forge/linux-64::libgcc-ng-12.2.0-h65d4601_19
libgfortran-ng conda-forge/linux-64::libgfortran-ng-12.2.0-h69a702a_19
libgfortran5 conda-forge/linux-64::libgfortran5-12.2.0-h337968e_19
libgomp conda-forge/linux-64::libgomp-12.2.0-h65d4601_19
liblapack conda-forge/linux-64::liblapack-3.9.0-16_linux64_openblas
libnsl conda-forge/linux-64::libnsl-2.0.0-h7f98852_0
libopenblas conda-forge/linux-64::libopenblas-0.3.21-pthreads_h78a6416_3
libsqlite conda-forge/linux-64::libsqlite-3.40.0-h753d276_0
libstdcxx-ng conda-forge/linux-64::libstdcxx-ng-12.2.0-h46fd767_19
libuuid conda-forge/linux-64::libuuid-2.32.1-h7f98852_1000
libzlib conda-forge/linux-64::libzlib-1.2.13-h166bdaf_4
ncurses conda-forge/linux-64::ncurses-6.3-h27087fc_1
numpy conda-forge/linux-64::numpy-1.24.2-py311h8e6699e_0
openssl conda-forge/linux-64::openssl-3.0.7-h0b41bf4_2
pip conda-forge/noarch::pip-23.0-pyhd8ed1ab_0
plasmidfinder bioconda/noarch::plasmidfinder-2.1.6-py310hdfd78af_1
python conda-forge/linux-64::python-3.11.0-he550d4f_1_cpython
python_abi conda-forge/linux-64::python_abi-3.11-3_cp311
readline conda-forge/linux-64::readline-8.1.2-h0f457ee_0
setuptools conda-forge/noarch::setuptools-67.1.0-pyhd8ed1ab_0
tabulate conda-forge/noarch::tabulate-0.9.0-pyhd8ed1ab_1
tk conda-forge/linux-64::tk-8.6.12-h27826a3_0
tzdata conda-forge/noarch::tzdata-2022g-h191b570_0
wheel conda-forge/noarch::wheel-0.38.4-pyhd8ed1ab_0
xz conda-forge/linux-64::xz-5.2.6-h166bdaf_0
zlib conda-forge/linux-64::zlib-1.2.13-h166bdaf_4
zstd conda-forge/linux-64::zstd-1.5.2-h3eb15da_6
Proceed ([y]/n)? y
Downloading and Extracting Packages
CondaError: Downloaded bytes did not match Content-Length
url: https://conda.anaconda.org/conda-forge/linux-64/numpy-1.24.2-py311h8e6699e_0.conda
target_path: /share/app/miniconda3/pkgs/numpy-1.24.2-py311h8e6699e_0.conda
Content-Length: 7720502
downloaded bytes: 6650303
CondaHTTPError: HTTP 000 CONNECTION FAILED for url <https://conda.anaconda.org/conda-forge/linux-64/boost-
cpp-1.80.0-h75c5d50_0.tar.bz2>Elapsed: -
An HTTP error occurred when trying to retrieve this URL.
HTTP errors are often intermittent, and a simple retry will get you on your way.
(plasmidfinder) root@mgt:/share/app/miniconda3/envs/arg/conda-meta# conda install -c bioconda -c conda-for
ge plasmidfinderCollecting package metadata (current_repodata.json): failed
# >>>>>>>>>>>>>>>>>>>>>> ERROR REPORT <<<<<<<<<<<<<<<<<<<<<<
Traceback (most recent call last):
File "/share/app/miniconda3/lib/python3.10/site-packages/urllib3/response.py", line 761, in _update_
chunk_length self.chunk_left = int(line, 16)
ValueError: invalid literal for int() with base 16: b''
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/share/app/miniconda3/lib/python3.10/site-packages/urllib3/response.py", line 444, in _error_c
atcher yield
File "/share/app/miniconda3/lib/python3.10/site-packages/urllib3/response.py", line 828, in read_chu
nked self._update_chunk_length()
File "/share/app/miniconda3/lib/python3.10/site-packages/urllib3/response.py", line 765, in _update_
chunk_length raise InvalidChunkLength(self, line)
urllib3.exceptions.InvalidChunkLength: InvalidChunkLength(got length b'', 0 bytes read)
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/share/app/miniconda3/lib/python3.10/site-packages/requests/models.py", line 816, in generate
yield from self.raw.stream(chunk_size, decode_content=True)
File "/share/app/miniconda3/lib/python3.10/site-packages/urllib3/response.py", line 624, in stream
for line in self.read_chunked(amt, decode_content=decode_content):
File "/share/app/miniconda3/lib/python3.10/site-packages/urllib3/response.py", line 816, in read_chu
nked with self._error_catcher():
File "/share/app/miniconda3/lib/python3.10/contextlib.py", line 153, in __exit__
self.gen.throw(typ, value, traceback)
File "/share/app/miniconda3/lib/python3.10/site-packages/urllib3/response.py", line 461, in _error_c
atcher raise ProtocolError("Connection broken: %r" % e, e)
urllib3.exceptions.ProtocolError: ("Connection broken: InvalidChunkLength(got length b'', 0 bytes read
)", InvalidChunkLength(got length b'', 0 bytes read))
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/exceptions.py", line 1118, in __call_
_ return func(*args, **kwargs)
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/cli/main.py", line 69, in main_subshe
ll exit_code = do_call(args, p)
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/cli/conda_argparse.py", line 91, in d
o_call return getattr(module, func_name)(args, parser)
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/notices/core.py", line 109, in wrappe
r return func(*args, **kwargs)
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/cli/main_install.py", line 20, in exe
cute install(args, parser, 'install')
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/cli/install.py", line 264, in install
unlink_link_transaction = solver.solve_for_transaction(
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/core/solve.py", line 136, in solve_fo
r_transaction unlink_precs, link_precs = self.solve_for_diff(update_modifier, deps_modifier,
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/core/solve.py", line 179, in solve_fo
r_diff final_precs = self.solve_final_state(update_modifier, deps_modifier, prune, ignore_pinned,
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/core/solve.py", line 284, in solve_fi
nal_state ssc = self._collect_all_metadata(ssc)
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/common/io.py", line 84, in decorated
return f(*args, **kwds)
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/core/solve.py", line 451, in _collect
_all_metadata index, r = self._prepare(prepared_specs)
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/core/solve.py", line 1061, in _prepar
e reduced_index = get_reduced_index(self.prefix, self.channels,
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/core/index.py", line 272, in get_redu
ced_index new_records = SubdirData.query_all(spec, channels=channels, subdirs=subdirs,
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/core/subdir_data.py", line 142, in qu
ery_all result = tuple(concat(executor.map(subdir_query, channel_urls)))
File "/share/app/miniconda3/lib/python3.10/concurrent/futures/_base.py", line 621, in result_iterato
r yield _result_or_cancel(fs.pop())
File "/share/app/miniconda3/lib/python3.10/concurrent/futures/_base.py", line 319, in _result_or_can
cel return fut.result(timeout)
File "/share/app/miniconda3/lib/python3.10/concurrent/futures/_base.py", line 458, in result
return self.__get_result()
File "/share/app/miniconda3/lib/python3.10/concurrent/futures/_base.py", line 403, in __get_result
raise self._exception
File "/share/app/miniconda3/lib/python3.10/concurrent/futures/thread.py", line 58, in run
result = self.fn(*self.args, **self.kwargs)
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/core/subdir_data.py", line 131, in <l
ambda> subdir_query = lambda url: tuple(
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/core/subdir_data.py", line 147, in qu
ery self.load()
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/core/subdir_data.py", line 213, in lo
ad _internal_state = self._load()
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/core/subdir_data.py", line 303, in _l
oad raw_repodata_str = fetch_repodata_remote_request(
File "/share/app/miniconda3/lib/python3.10/site-packages/conda/core/subdir_data.py", line 571, in fe
tch_repodata_remote_request resp = session.get(
File "/share/app/miniconda3/lib/python3.10/site-packages/requests/sessions.py", line 600, in get
return self.request("GET", url, **kwargs)
File "/share/app/miniconda3/lib/python3.10/site-packages/requests/sessions.py", line 587, in request
resp = self.send(prep, **send_kwargs)
File "/share/app/miniconda3/lib/python3.10/site-packages/requests/sessions.py", line 745, in send
r.content
File "/share/app/miniconda3/lib/python3.10/site-packages/requests/models.py", line 899, in content
self._content = b"".join(self.iter_content(CONTENT_CHUNK_SIZE)) or b""
File "/share/app/miniconda3/lib/python3.10/site-packages/requests/models.py", line 818, in generate
raise ChunkedEncodingError(e)
requests.exceptions.ChunkedEncodingError: ("Connection broken: InvalidChunkLength(got length b'', 0 by
tes read)", InvalidChunkLength(got length b'', 0 bytes read))
`$ /share/app/miniconda3/bin/conda install -c bioconda -c conda-forge plasmidfinder`
environment variables:
CIO_TEST=<not set>
CONDA_DEFAULT_ENV=plasmidfinder
CONDA_EXE=/share/app/miniconda3/bin/conda
CONDA_PREFIX=/share/app/miniconda3/envs/plasmidfinder
CONDA_PREFIX_1=/share/app/miniconda3
CONDA_PREFIX_2=/share/app/miniconda3/envs/motus
CONDA_PREFIX_3=/share/app/miniconda3/envs/phyloflash
CONDA_PREFIX_4=/share/app/miniconda3
CONDA_PREFIX_5=/share/app/miniconda3/envs/plasflow
CONDA_PROMPT_MODIFIER=(plasmidfinder)
CONDA_PYTHON_EXE=/share/app/miniconda3/bin/python
CONDA_ROOT=/share/app/miniconda3
CONDA_SHLVL=6
CURL_CA_BUNDLE=<not set>
JAVA_LD_LIBRARY_PATH=:-:-:-:-:-:-:-
LD_LIBRARY_PATH=/public/arg/ISEScan-1.7.2.3:/public/arg/ISEScan-1.7.2.3:
LD_PRELOAD=<not set>
MANPATH=/opt/xcat/share/man::/usr/bin/ypuser/man
MODULEPATH=/share/modulefiles:/root/modulefiles:/usr/share/Modules/modulefiles:/s
hare/app/oneapi2022/modulefiles
PATH=/public/arg/phyloFlash-
pf3.4:/public/arg/FragGeneScan:/public/nanoplot/ncbi-blast-
2.13.0+/bin:/public/nanoplot/centrifuge-1.0.3-
beta:/public/nanoplot/sratoolkit.2.10.8-
centos_linux64/bin:/public/metagenome/microbiome_helper-master:/share/
app/miniconda3/envs/plasmidfinder/bin:/public/arg/FragGeneScan:/public
/nanoplot/ncbi-blast-2.13.0+/bin:/public/nanoplot/centrifuge-1.0.3-
beta:/public/nanoplot/sratoolkit.2.10.8-
centos_linux64/bin:/public/metagenome/microbiome_helper-master:/share/
app/miniconda3/bin:/share/app/miniconda3/condabin:/share/app/miniconda
3/bin:/opt/xcat/bin:/opt/xcat/sbin:/opt/xcat/share/xcat/tools:/usr/lib
64/qt-
3.3/bin:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/usr/bin/ypu
ser:/root/bin
REQUESTS_CA_BUNDLE=<not set>
SSL_CERT_FILE=<not set>
active environment : plasmidfinder
active env location : /share/app/miniconda3/envs/plasmidfinder
shell level : 6
user config file : /root/.condarc
populated config files :
conda version : 22.11.1
conda-build version : not installed
python version : 3.10.9.final.0
virtual packages : __archspec=1=x86_64
__glibc=2.17=0
__linux=3.10.0=0
__unix=0=0
base environment : /share/app/miniconda3 (writable)
conda av data dir : /share/app/miniconda3/etc/conda
conda av metadata url : None
channel URLs : https://conda.anaconda.org/bioconda/linux-64
https://conda.anaconda.org/bioconda/noarch
https://conda.anaconda.org/conda-forge/linux-64
https://conda.anaconda.org/conda-forge/noarch
https://repo.anaconda.com/pkgs/main/linux-64
https://repo.anaconda.com/pkgs/main/noarch
https://repo.anaconda.com/pkgs/r/linux-64
https://repo.anaconda.com/pkgs/r/noarch
package cache : /share/app/miniconda3/pkgs
/root/.conda/pkgs
envs directories : /share/app/miniconda3/envs
/root/.conda/envs
platform : linux-64
user-agent : conda/22.11.1 requests/2.28.1 CPython/3.10.9 Linux/3.10.0-1160.el7.x86_64 centos
/7.9.2009 glibc/2.17 UID:GID : 0:0
netrc file : None
offline mode : False
An unexpected error has occurred. Conda has prepared the above report.
If submitted, this report will be used by core maintainers to improve
future releases of conda.
Would you like conda to send this report to the core maintainers? [y/N]:
Timeout reached. No report sent.
virsorter
"Collecting package metadata (current_repodata.json): done" 表示 conda 正在收集当前存储库中的软件包元数据。元数据包括软件包的版本,依赖关系等信息,conda 需要这些信息来确定如何安装该软件包。
"Solving environment: failed with initial frozen solve. Retrying with flexible solve." 意味着 conda 在初始冻结的解决方案中失败了,因此在使用更加灵活的解决方案时再次尝试安装。这种情况通常表示与当前环境中的软件包冲突,导致 conda 无法安装 mamba。
在这种情况下,建议您创建一个新的 conda 环境,并在该环境中安装该软件包,以避免与当前环境中的软件包冲突。您可以使用以下命令创建新环境:Option 2 (development version)
The development version is most updated. To install the development version:
conda create -n vs2 -c conda-forge -c bioconda "python>=3.6" scikit-learn=0.22.1 imbalanced-learn pandas seaborn hmmer==3.3 prodigal screed ruamel.yaml "snakemake>=5.18,<=5.26" click mamba conda activate vs2 git clone https://github.com/jiarong/VirSorter2.git cd VirSorter2 pip install -e .
apptainer singularity centos系统
apptainer/INSTALL.md at main · apptainer/apptainer · GitHub
CentOS 7安装Development Tools失败 – 简书
sudo yum groupinstall -y 'Development Tools'On CentOS/RHEL:
# Install basic tools for compiling
sudo yum groupinstall -y 'Development Tools'
# Ensure EPEL repository is available
sudo yum install -y epel-release
# Install RPM packages for dependencies
sudo yum install -y
libseccomp-devel
squashfs-tools
squashfuse
fuse-overlayfs
fakeroot
/usr/*bin/fuse2fs
cryptsetup
wget gitDownloads – The Go Programming Language
udo tar -C /usr/local -xzf /tmp/go1.20.linux-amd64.tar.gz 什么意思
The sudo tar -C /usr/local -xzf /tmp/go1.20.linux-amd64.tar.gz command has the following meaning:
sudo: The command is executed with superuser (root) privileges.
tar: The tar (tape archive) command is used to extract files from a tar archive.
-C /usr/local: The -C option specifies the directory (/usr/local) where the tar archive should be extracted.
-xzf: The -x option extracts the files, the -z option uncompresses the archive using gzip, and the -f option specifies the archive file name (/tmp/go1.20.linux-amd64.tar.gz).
So, the overall effect of the command is to extract the contents of the /tmp/go1.20.linux-amd64.tar.gz archive to the /usr/local directory.git clone —recursive https://github.com/apptainer/apptainer.git 需要加recursive比较稳妥
Clone the repo
With the adoption of Go modules you no longer need to clone the Apptainer repository to a specific location.
Clone the repository with git in a location of your choice:
git clone https://github.com/apptainer/apptainer.git
cd apptainer
By default your clone will be on the main branch which is where development of Apptainer happens. To build a specific version of Apptainer, check out a release tag before compiling, for example:
git checkout v1.1.5
Compiling Apptainer
You can configure, build, and install Apptainer using the following commands:
./mconfig
cd ./builddir
make
sudo make install(Linux)make编译用法简述_漠效的博客-CSDN博客_make编译
【1】make
等同于 make all
它是一个解释makefile中指令的命令工具
执行make时,默认会去(当前文件夹下),找名为Makefile的文件
根据Makefile文件来编译源代码,生成目标文件及可执行文件
在每次使用make时,都会重新生成objects,新产生的object会覆盖旧的objects
【2】make clean
清除以前的make命令编译后所产生的object文件(即后缀为“.o”的文件)及其生成的可执行文件
一般来说,在每次使用make时,都会重新生成objects,新产生的object会覆盖旧的objects
但是实际用时,经过多次编译后,必须使用make clean(直接make,以前留下来的错误不能清除干净)
【3】make install
将编译成功的可执行文件移动到make编译时填写的目录,依据生成的makefile文件
一般默认系统目录/usr/local/bin
【4】make dist
产生发布软件包.
即将可执行文件及相关文件打包成一个tar.gz格式的压缩文件,会在当前目录下生成XXX.tar.gz的文件
【5】make distcheck
相当于make dist命令的进化版
生成发布软件包并对其进行检查,以确定发布包的正确性
它会自动把压缩包文件解开,然后执行configure命令,并且执行make,来确认编译不出现错误,并会在最后提示你软件包是否正确
【6】make distclean
类似于make clean
清除以前的make命令编译后所产生的object文件(即后缀为“.o”的文件)及其生成的可执行文件,并将configure生成的文件全部删除掉
通常在使用make clean后使用,清理的更干净,避免影响再次编译
makefile概念
makefile有些类似于脚本,通过定义了一系列的规则,指定哪些文件需要先编译,哪些文件需要后编译,哪些文件需要重新编译,甚至于进行更复杂的功能操作,并且它也可以执行操作系统的命令
当编译安装源码包时,使用make编译,它会默认找当前目录下的makefile文件,依据makefile中的规则,执行编译安装
makefile可以实现自动化编译,原理上来说,只需要一个make命令,整个工程完全自动编译,极大的提高了软件开发的效率
makefile成为了一种被广泛应用于在工程方面的编译方法
————————————————
版权声明:本文为CSDN博主「漠效」的原创文章,遵循CC 4.0 BY-SA版权协议,转载请附上原文出处链接及本声明。
原文链接:https://blog.csdn.net/gx_1_11_real/article/details/81068412at Makefile # # Non-recursive Makefile GENERATED by `mconfig' -- Wed Feb 8 02:10:40 CST 2023 # configured: /public/arg/apptainer/mconfig --with-suid # # [ Embedding file : /public/arg/apptainer/builddir/makeit-intermediate/base.mk ] # # silent build V := @ # source/build locations BUILDDIR := . BUILDDIR_ABSPATH := /public/arg/apptainer/builddir SOURCEDIR := /public/arg/apptainer SCRIPTSDIR := /public/arg/apptainer/mlocal/scripts CONTRIBDIR := PREFIX := /usr/local EXECPREFIX := /usr/local BINDIR := /usr/local/bin SBINDIR := /usr/local/sbin LIBEXECDIR := /usr/local/libexec DATAROOTDIR := /usr/local/share DATADIR := /usr/local/share SYSCONFDIR := /usr/local/etc SHAREDSTATEDIR := /usr/local/com LOCALSTATEDIR := /usr/local/var RUNSTATEDIR := /usr/local/var/run INCLUDEDIR := /usr/local/include OLDINCLUDEDIR := /usr/include DOCDIR := /usr/local/share/doc/apptainer INFODIR := /usr/local/share/info HTMLDIR := /usr/local/share/doc/apptainer DVIDIR := /usr/local/share/doc/apptainer PDFDIR := /usr/local/share/doc/apptainer PSDIR := /usr/local/share/doc/apptainer LIBDIR := /usr/local/lib LOCALEDIR := /usr/local/share/locale MANDIR := /usr/local/share/man RPMPREFIX:= NAME := apptainer SHORT_VERSION := 1.1.5 VERSION := 1.1.5 HOSTAR := ar HOSTCC := cc HOSTCXX := c++ HOSTLD := cc HOSTRANLIB := ranlib HOSTSIZE := size HOSTOBJCOPY := objcopy AR := ar CC := cc CXX := c++ LD := ld RANLIB := ranlib SIZE := size OBJCOPY := objcopy ARCH := x86_64 CFLAGS := -D_FORTIFY_SOURCE=2 -Wformat -Wformat-security -fstack-protector --param ssp-buffer-size=4 -Wal l -Werror -Wfatal-errors -Wno-unknown-warning-option -Wstrict-prototypes -Wpointer-arith -Wbad-function-cast -Woverlength-strings -Wframe-larger-than=2047 -Wno-sign-compare -Wclobbered -Wempty-body -Wmissing-parameter-type -Wtype-limits -Wunused-parameter -Wunused-but-set-parameter -Wno-discarded-qualifiers -Wno-incompatible-pointer-types -pipe -fmessage-length=0 -fPIC GO := /usr/local/go/bin/go CGO_CFLAGS := -I/public/arg/apptainer/builddir CGO_LDFLAGS := CGO_CPPFLAGS := CGO_CXXFLAGS := export CGO_CFLAGS CGO_LDFLAGS CGO_CPPFLAGS CGO_CXXFLAGS LDFLAGS := # make sure `all' is the first rule all: # [ Embedding file : /public/arg/apptainer/mlocal/frags/common_opts.mk ] # # a list of extra files to clean augmented by each module (*.mconf) file CLEANFILES := # general build-wide compile options AFLAGS := -g LDFLAGS += -Wl,-z,relro,-z,now # [ Embedding file : /public/arg/apptainer/mlocal/frags/go_common_opts.mk ] # # go tool default build options GO111MODULE := on GO_TAGS := containers_image_openpgp sylog oci_engine apptainer_engine fakeroot_engine GO_TAGS_SUID := containers_image_openpgp sylog apptainer_engine fakeroot_engine GO_LDFLAGS := # Need to use non-pie build on ppc64le # https://github.com/apptainer/singularity/issues/5762 # Need to disable race detector on ppc64le # https://github.com/apptainer/singularity/issues/5914 uname_m := $(shell uname -m) ifeq ($(uname_m),ppc64le) GO_BUILDMODE := -buildmode=default GO_RACE := else GO_BUILDMODE := -buildmode=pie GO_RACE := -race endif GO_MODFLAGS := $(if $(wildcard $(SOURCEDIR)/vendor/modules.txt),-mod=vendor,-mod=readonly) GO_MODFILES := $(SOURCEDIR)/go.mod $(SOURCEDIR)/go.sum GOFLAGS := $(GO_MODFLAGS) -trimpath GOPROXY := https://proxy.golang.org export GOFLAGS GO111MODULE GOPROXY # [ Embedding file : /public/arg/apptainer/mlocal/frags/go_unix_opts.mk ] # GO_TAGS += apparmor selinux GO_TAGS_SUID += apparmor selinux # [ Embedding file : /public/arg/apptainer/mlocal/frags/go_runtime_opts.mk ] # CGO_CFLAGS += -O2 -D_FORTIFY_SOURCE=2 -Wformat -Wformat-security -fstack-protector --param ssp-buffer-size =4 # [ Embedding file : /public/arg/apptainer/mlocal/frags/release_opts.mk ] # CFLAGS += -O2 LDFLAGS += -s # [ Embedding file : /public/arg/apptainer/mlocal/frags/go_normal_opts.mk ] # # This tells go's link command to add a GNU Build Id, needed for later # symbol stripping for example as is done by rpmbuild. GO_LDFLAGS += -ldflags="-B 0x`head -c20 /dev/urandom|od -An -tx1|tr -d ' n'`" # [ Embedding file : /public/arg/apptainer/mlocal/frags/arch_x86_64_opts.mk ] # # [ Embedding file : /public/arg/apptainer/builddir/makeit-intermediate/combined-mconfsready.mk ] # # object files list _OBJ := # suffix rules (metarules missing from most variants) # [ Embedding file : /public/arg/apptainer/builddir/makeit-intermediate/all.mk ] # all: $() CLEANFILES += # [ Embedding file : /public/arg/apptainer/mlocal/frags/build_cli.mk ] # # This file contains all of the rules for building the apptainer CLI binary # apptainer build config apptainer_build_config := $(SOURCEDIR)/internal/pkg/buildcfg/config.go $(apptainer_build_config): $(BUILDDIR)/config.h $(SOURCEDIR)/scripts/go-generate $(V)rm -f $(apptainer_build_config) $(V) cd $(SOURCEDIR)/internal/pkg/buildcfg && $(SOURCEDIR)/scripts/go-generate CLEANFILES += $(apptainer_build_config) # contain apptainer_SOURCE variable list apptainer_deps := $(BUILDDIR_ABSPATH)/apptainer.deps -include $(apptainer_deps) $(apptainer_deps): $(GO_MODFILES) @echo " GEN GO DEP" $@ $(V)$(SOURCEDIR)/makeit/gengodep -v3 "$(GO)" "apptainer_SOURCE" "$(GO_TAGS)" "$@" "$(SOURCEDIR)/cmd /apptainer" # Look at dependencies file changes via apptainer_deps # because it means that a module was updated. apptainer := $(BUILDDIR)/apptainer $(apptainer): $(apptainer_build_config) $(apptainer_deps) $(apptainer_SOURCE) @echo " GO" $@; echo " [+] GO_TAGS" "$(GO_TAGS)" $(V)$(GO) build $(GO_MODFLAGS) $(GO_BUILDMODE) -tags "$(GO_TAGS)" $(GO_LDFLAGS) -o $(BUILDDIR)/apptainer $(SOURCEDIR)/cmd/apptainer apptainer_INSTALL := $(DESTDIR)$(BINDIR)/apptainer $(apptainer_INSTALL): $(apptainer) @echo " INSTALL" $@ $(V)umask 0022 && mkdir -p $(@D) $(V)install -m 0755 $(apptainer) $(apptainer_INSTALL) # set cp to install singularity_INSTALL := $(DESTDIR)$(BINDIR)/singularity $(singularity_INSTALL): @echo " INSTALL" $@ $(V)ln -sf apptainer $(singularity_INSTALL) CLEANFILES += $(apptainer) INSTALLFILES += $(apptainer_INSTALL) $(singularity_INSTALL) ALL += $(apptainer) # bash_completion files bash_completion1 := $(BUILDDIR)/etc/bash_completion.d/apptainer bash_completion2 := $(BUILDDIR)/etc/bash_completion.d/singularity bash_completions := $(bash_completion1) $(bash_completion2) $(bash_completions): $(apptainer_build_config) @echo " GEN" $@ $(V)rm -f $@ $(V)mkdir -p $(@D) $(V)$(GO) run $(GO_MODFLAGS) -tags "$(GO_TAGS)" $(SOURCEDIR)/cmd/bash_completion/bash_completion.go $@ bash_completion1_INSTALL := $(DESTDIR)$(DATADIR)/bash-completion/completions/apptainer bash_completion2_INSTALL := $(DESTDIR)$(DATADIR)/bash-completion/completions/singularity bash_completions_INSTALL := $(bash_completion1_INSTALL) $(bash_completion2_INSTALL) $(bash_completion1_INSTALL): $(bash_completion1) @echo " INSTALL" $@ $(V)umask 0022 && mkdir -p $(@D) $(V)install -m 0644 $< $@ $(bash_completion2_INSTALL): $(bash_completion2) @echo " INSTALL" $@ $(V)umask 0022 && mkdir -p $(@D) $(V)install -m 0644 $< $@ CLEANFILES += $(bash_completions) INSTALLFILES += $(bash_completions_INSTALL) ALL += $(bash_completions) # apptainer.conf file config := $(BUILDDIR)/apptainer.conf config_INSTALL := $(DESTDIR)$(SYSCONFDIR)/apptainer/apptainer.conf # override this to empty to avoid merging old configuration settings old_config := $(config_INSTALL) $(config): $(apptainer) @echo " GEN $@`if [ -n "$(old_config)" ]; then echo " from $(old_config)"; fi`" $(V)$(apptainer) confgen $(old_config) $(config) $(config_INSTALL): $(config) @echo " INSTALL" $@ $(V)umask 0022 && mkdir -p $(@D) $(V)install -m 0644 $< $@ INSTALLFILES += $(config_INSTALL) ALL += $(config) # remote config file remote_config := $(SOURCEDIR)/etc/remote.yaml remote_config_INSTALL := $(DESTDIR)$(SYSCONFDIR)/apptainer/remote.yaml $(remote_config_INSTALL): $(remote_config) @echo " INSTALL" $@ $(V)umask 0022 && mkdir -p $(@D) $(V)install -m 0644 $< $@ INSTALLFILES += $(remote_config_INSTALL) man_pages := $(BUILDDIR)$(MANDIR)/man1 $(man_pages): apptainer @echo " MAN" $@ mkdir -p $@ $(V)$(GO) run $(GO_MODFLAGS) -tags "$(GO_TAGS)" $(SOURCEDIR)/cmd/docs/docs.go man --dir $@ $(V)cd $@; for M in apptainer*; do S="`echo $$M|sed 's/apptainer/singularity/'`"; ln -fs $$M $$S; done man_pages_INSTALL := $(DESTDIR)$(MANDIR)/man1 $(man_pages_INSTALL): $(man_pages) @echo " INSTALL" $@ $(V)umask 0022 && mkdir -p $@ $(V)install -m 0644 -t $@ $(man_pages)/* INSTALLFILES += $(man_pages_INSTALL) ALL += $(man_pages) # [ Embedding file : /public/arg/apptainer/mlocal/frags/build_network.mk ] # # This file contains rules for building CNI plugins which can enable # different networking functions between container(s) and the host apptainer_REPO := github.com/apptainer/apptainer cni_builddir := $(BUILDDIR_ABSPATH)/cni cni_install_DIR := $(DESTDIR)$(LIBEXECDIR)/apptainer/cni cni_plugins := $(shell grep '^ _' $(SOURCEDIR)/internal/pkg/runtime/engine/apptainer/plugins_linux.go | c ut -d" -f2)cni_plugins_EXECUTABLES := $(addprefix $(cni_builddir)/, $(notdir $(cni_plugins))) cni_plugins_INSTALL := $(addprefix $(cni_install_DIR)/, $(notdir $(cni_plugins))) cni_config_LIST := $(SOURCEDIR)/etc/network/00_bridge.conflist $(SOURCEDIR)/etc/network/10_ptp.conflist $(SOURCEDIR)/etc/network/20_ipvlan.conflist $(SOURCEDIR)/etc/network/30_macvlan.conflist $(SOURCEDIR)/etc/network/40_fakeroot.conflist cni_config_INSTALL := $(DESTDIR)$(SYSCONFDIR)/apptainer/network .PHONY: cniplugins cniplugins: $(V)umask 0022 && mkdir -p $(cni_builddir) $(V)for p in $(cni_plugins); do name=`basename $$p`; cniplugin=$(cni_builddir)/$$name; if [ ! -f $$cniplugin ]; then echo " CNI PLUGIN" $$name; $(GO) build $(GO_MODFLAGS) $(GO_BUILDMODE) -tags "$(GO_TAGS)" $(GO_LDFLAGS) -o $$cniplugin $$p; fi done# run-singularity script
run_singularity := $(SOURCEDIR)/scripts/run-singularityrun_singularity_INSTALL := $(DESTDIR)$(BINDIR)/run-singularity
$(run_singularity_INSTALL): $(run_singularity)
@echo ” INSTALL” $@
$(V)umask 0022 && mkdir -p $(@D)
$(V)install -m 0755 $< $@INSTALLFILES += $(run_singularity_INSTALL)
# capability config file
capability_config_INSTALL := $(DESTDIR)$(SYSCONFDIR)/apptainer/capability.json
$(capability_config_INSTALL):
@echo ” INSTALL” $@
$(V)umask 0022 && mkdir -p $(@D)
$(V)touch $@INSTALLFILES += $(capability_config_INSTALL)
go env GOPATH
make编译过程中用到的或者下载下来的文件在哪 go下载的临时文件都在哪里呢 通常在go env GOPATH 路径下面
ls -lha /root/go/pkg/m
od/cache/download/github.com/total 4.0K
drwxr-xr-x 25 root root 4.0K Feb 8 02:14 .
drwxr-xr-x 7 root root 91 Feb 8 02:13 ..
drwxr-xr-x 3 root root 17 Feb 8 02:14 apex
drwxr-xr-x 5 root root 77 Feb 8 02:13 apptainer
drwxr-xr-x 3 root root 20 Feb 8 02:11 blang
drwxr-xr-x 3 root root 24 Feb 8 02:13 containerd
drwxr-xr-x 4 root root 32 Feb 8 02:11 containernetworking
drwxr-xr-x 3 root root 19 Feb 8 02:11 containers
drwxr-xr-x 3 root root 17 Feb 8 02:11 creack
drwxr-xr-x 3 root root 33 Feb 8 02:13 cyphar
drwxr-xr-x 4 root root 36 Feb 8 02:14 docker
drwxr-xr-x 3 root root 19 Feb 8 02:13 fatih
drwxr-xr-x 3 root root 17 Feb 8 02:14 go-log
drwxr-xr-x 3 root root 18 Feb 8 02:11 google
drwxr-xr-x 3 root root 18 Feb 8 02:14 gosimple
drwxr-xr-x 3 root root 27 Feb 8 02:11 hashicorp
drwxr-xr-x 8 root root 101 Feb 8 02:14 opencontainers
drwxr-xr-x 3 root root 21 Feb 8 02:11 pelletier
drwxr-xr-x 3 root root 20 Feb 8 02:11 pkg
drwxr-xr-x 3 root root 23 Feb 8 02:11 !proton!mail
drwxr-xr-x 3 root root 31 Feb 8 02:11 seccomp
drwxr-xr-x 3 root root 21 Feb 8 02:13 shopspring
drwxr-xr-x 4 root root 32 Feb 8 02:13 spf13
drwxr-xr-x 3 root root 23 Feb 8 02:13 sylabs
drwxr-xr-x 3 root root 17 Feb 8 02:13 vbauerster
make
GO apptainer
[+] GO_TAGS "containers_image_openpgp sylog oci_engine apptainer_engine fakeroot_engine apparmor selin
ux"go: downloading github.com/apptainer/container-key-client v0.8.0
go: downloading github.com/apptainer/sif/v2 v2.8.1
go: downloading github.com/fatih/color v1.13.0
go: downloading golang.org/x/sys v0.0.0-20220715151400-c0bba94af5f8
go: downloading github.com/apptainer/container-library-client v1.3.3
go: downloading github.com/sylabs/json-resp v0.8.1
go: downloading github.com/creack/pty v1.1.18
go: downloading github.com/opencontainers/runtime-spec v1.0.3-0.20210326190908-1c3f411f0417
go: downloading github.com/shopspring/decimal v1.3.1
go: downloading github.com/ProtonMail/go-crypto v0.0.0-20220824120805-4b6e5c587895
go: downloading gopkg.in/yaml.v2 v2.4.0
go: downloading github.com/docker/go-units v0.4.0
go: downloading golang.org/x/term v0.0.0-20210927222741-03fcf44c2211
go: downloading github.com/spf13/cobra v1.5.0
go: downloading github.com/containers/image/v5 v5.22.0
go: downloading github.com/spf13/pflag v1.0.5
go: downloading github.com/seccomp/containers-golang v0.6.0
go: downloading github.com/containerd/containerd v1.6.8
go: downloading github.com/blang/semver/v4 v4.0.0
go: downloading github.com/pkg/errors v0.9.1
go: downloading github.com/pelletier/go-toml v1.9.5
go: downloading github.com/opencontainers/runc v1.1.4
go: downloading github.com/opencontainers/go-digest v1.0.0
go: downloading github.com/opencontainers/image-spec v1.0.2-0.20211117181255-693428a734f5
go: downloading oras.land/oras-go v1.2.0
go: downloading github.com/go-log/log v0.2.0
go: downloading github.com/gosimple/slug v1.12.0
go: downloading github.com/vbauerster/mpb/v7 v7.4.2
go: downloading mvdan.cc/sh/v3 v3.5.1
go: downloading github.com/cyphar/filepath-securejoin v0.2.3
go: downloading github.com/google/uuid v1.3.0
go: downloading github.com/apex/log v1.9.0
go: downloading github.com/opencontainers/umoci v0.4.7
go: downloading github.com/docker/docker v20.10.17+incompatible
go: downloading github.com/opencontainers/selinux v1.10.1
../cmd/internal/cli/pgp.go:23:2: github.com/ProtonMail/go-crypto@v0.0.0-20220824120805-4b6e5c587895: Get "
https://proxy.golang.org/github.com/%21proton%21mail/go-crypto/@v/v0.0.0-20220824120805-4b6e5c587895.zip": dial tcp 172.217.160.113:443: i/o timeout../pkg/sypgp/sypgp.go:35:2: github.com/ProtonMail/go-crypto@v0.0.0-20220824120805-4b6e5c587895: Get "https
://proxy.golang.org/github.com/%21proton%21mail/go-crypto/@v/v0.0.0-20220824120805-4b6e5c587895.zip": dial tcp 172.217.160.113:443: i/o timeout../cmd/internal/cli/pgp.go:24:2: github.com/ProtonMail/go-crypto@v0.0.0-20220824120805-4b6e5c587895: Get "
https://proxy.golang.org/github.com/%21proton%21mail/go-crypto/@v/v0.0.0-20220824120805-4b6e5c587895.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/sources/oci_unpack.go:23:2: github.com/apex/log@v1.9.0: Get "https://proxy.golang.or
g/github.com/apex/log/@v/v1.9.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/remote/endpoint/client.go:19:2: github.com/apptainer/container-key-client@v0.8.0: Get "htt
ps://proxy.golang.org/github.com/apptainer/container-key-client/@v/v0.8.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/remote/endpoint/client.go:20:2: github.com/apptainer/container-library-client@v1.3.3: Get
"https://proxy.golang.org/github.com/apptainer/container-library-client/@v/v1.3.3.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/app/apptainer/sign.go:14:2: github.com/apptainer/sif/v2@v2.8.1: Get "https://proxy.golang.org/
github.com/apptainer/sif/v2/@v/v2.8.1.zip": dial tcp 172.217.160.113:443: i/o timeout../pkg/image/sif.go:19:2: github.com/apptainer/sif/v2@v2.8.1: Get "https://proxy.golang.org/github.com/app
tainer/sif/v2/@v/v2.8.1.zip": dial tcp 172.217.160.113:443: i/o timeout../cmd/internal/cli/siftool.go:15:2: github.com/apptainer/sif/v2@v2.8.1: Get "https://proxy.golang.org/git
hub.com/apptainer/sif/v2/@v/v2.8.1.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/plugin/module.go:23:2: github.com/blang/semver/v4@v4.0.0: Get "https://proxy.golang.org/gi
thub.com/blang/semver/v4/@v/v4.0.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/client/oras/oras.go:29:2: github.com/containerd/containerd@v1.6.8: Get "https://proxy.gola
ng.org/github.com/containerd/containerd/@v/v1.6.8.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/client/oras/oras.go:30:2: github.com/containerd/containerd@v1.6.8: Get "https://proxy.gola
ng.org/github.com/containerd/containerd/@v/v1.6.8.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/client/oras/oras.go:31:2: github.com/containerd/containerd@v1.6.8: Get "https://proxy.gola
ng.org/github.com/containerd/containerd/@v/v1.6.8.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/client/oras/oras.go:32:2: github.com/containerd/containerd@v1.6.8: Get "https://proxy.gola
ng.org/github.com/containerd/containerd/@v/v1.6.8.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/oci/oci.go:22:2: github.com/containers/image/v5@v5.22.0: Get "https://proxy.golang.o
rg/github.com/containers/image/v5/@v/v5.22.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/oci/oci.go:23:2: github.com/containers/image/v5@v5.22.0: Get "https://proxy.golang.o
rg/github.com/containers/image/v5/@v/v5.22.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/sources/conveyorPacker_oci.go:38:2: github.com/containers/image/v5@v5.22.0: Get "htt
ps://proxy.golang.org/github.com/containers/image/v5/@v/v5.22.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/sources/conveyorPacker_oci.go:39:2: github.com/containers/image/v5@v5.22.0: Get "htt
ps://proxy.golang.org/github.com/containers/image/v5/@v/v5.22.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/sources/conveyorPacker_oci.go:40:2: github.com/containers/image/v5@v5.22.0: Get "htt
ps://proxy.golang.org/github.com/containers/image/v5/@v/v5.22.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/oci/oci.go:24:2: github.com/containers/image/v5@v5.22.0: Get "https://proxy.golang.o
rg/github.com/containers/image/v5/@v/v5.22.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/oci/oci.go:25:2: github.com/containers/image/v5@v5.22.0: Get "https://proxy.golang.o
rg/github.com/containers/image/v5/@v/v5.22.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/oci/oci.go:26:2: github.com/containers/image/v5@v5.22.0: Get "https://proxy.golang.o
rg/github.com/containers/image/v5/@v/v5.22.0.zip": dial tcp 172.217.160.113:443: i/o timeout../pkg/build/types/bundle.go:24:2: github.com/containers/image/v5@v5.22.0: Get "https://proxy.golang.org/g
ithub.com/containers/image/v5/@v/v5.22.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/runtime/engine/oci/prepare_linux.go:24:2: github.com/creack/pty@v1.1.18: Get "https://prox
y.golang.org/github.com/creack/pty/@v/v1.1.18.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/files/files.go:19:2: github.com/cyphar/filepath-securejoin@v0.2.3: Get "https://prox
y.golang.org/github.com/cyphar/filepath-securejoin/@v/v0.2.3.zip": dial tcp 172.217.160.113:443: i/o timeout../pkg/util/archive/copy.go:17:2: github.com/docker/docker@v20.10.17+incompatible: Get "https://proxy.gola
ng.org/github.com/docker/docker/@v/v20.10.17+incompatible.zip": dial tcp 172.217.160.113:443: i/o timeout../pkg/util/archive/copy.go:18:2: github.com/docker/docker@v20.10.17+incompatible: Get "https://proxy.gola
ng.org/github.com/docker/docker/@v/v20.10.17+incompatible.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/app/apptainer/instance_linux.go:30:2: github.com/docker/go-units@v0.4.0: Get "https://proxy.go
lang.org/github.com/docker/go-units/@v/v0.4.0.zip": dial tcp 172.217.160.113:443: i/o timeout../cmd/internal/cli/pgp.go:31:2: github.com/fatih/color@v1.13.0: Get "https://proxy.golang.org/github.com/
fatih/color/@v/v1.13.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/remote/endpoint/client.go:21:2: github.com/go-log/log@v0.2.0: Get "https://proxy.golang.or
g/github.com/go-log/log/@v/v0.2.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/util/crypt/crypt_dev.go:28:2: github.com/google/uuid@v1.3.0: Get "https://proxy.golang.org
/github.com/google/uuid/@v/v1.3.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/remote/endpoint/config.go:24:2: github.com/gosimple/slug@v1.12.0: Get "https://proxy.golan
g.org/github.com/gosimple/slug/@v/v1.12.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/client/oras/oras.go:34:2: github.com/opencontainers/go-digest@v1.0.0: Get "https://proxy.g
olang.org/github.com/opencontainers/go-digest/@v/v1.0.0.zip": dial tcp 172.217.160.113:443: i/o timeout../pkg/ocibundle/sif/bundle_linux.go:21:2: github.com/opencontainers/image-spec@v1.0.2-0.20211117181255-69
3428a734f5: Get "https://proxy.golang.org/github.com/opencontainers/image-spec/@v/v1.0.2-0.20211117181255-693428a734f5.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/cgroups/manager_linux.go:22:2: github.com/opencontainers/runc@v1.1.4: Get "https://proxy.g
olang.org/github.com/opencontainers/runc/@v/v1.1.4.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/cgroups/manager_linux.go:24:2: github.com/opencontainers/runc@v1.1.4: Get "https://proxy.g
olang.org/github.com/opencontainers/runc/@v/v1.1.4.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/cgroups/manager_linux.go:25:2: github.com/opencontainers/runc@v1.1.4: Get "https://proxy.g
olang.org/github.com/opencontainers/runc/@v/v1.1.4.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/cgroups/manager_linux.go:26:2: github.com/opencontainers/runc@v1.1.4: Get "https://proxy.g
olang.org/github.com/opencontainers/runc/@v/v1.1.4.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/runtime/engine/config/oci/generate/generate.go:35:2: github.com/opencontainers/runtime-spe
c@v1.0.3-0.20210326190908-1c3f411f0417: Get "https://proxy.golang.org/github.com/opencontainers/runtime-spec/@v/v1.0.3-0.20210326190908-1c3f411f0417.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/security/selinux/selinux_supported.go:14:8: github.com/opencontainers/selinux@v1.10.1: Get
"https://proxy.golang.org/github.com/opencontainers/selinux/@v/v1.10.1.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/sources/oci_unpack.go:30:2: github.com/opencontainers/umoci@v0.4.7: Get "https://pro
xy.golang.org/github.com/opencontainers/umoci/@v/v0.4.7.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/sources/oci_unpack.go:31:2: github.com/opencontainers/umoci@v0.4.7: Get "https://pro
xy.golang.org/github.com/opencontainers/umoci/@v/v0.4.7.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/sources/oci_unpack.go:32:2: github.com/opencontainers/umoci@v0.4.7: Get "https://pro
xy.golang.org/github.com/opencontainers/umoci/@v/v0.4.7.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/cgroups/config_linux.go:18:2: github.com/pelletier/go-toml@v1.9.5: Get "https://proxy.gola
ng.org/github.com/pelletier/go-toml/@v/v1.9.5.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/util/bin/bin.go:22:2: github.com/pkg/errors@v0.9.1: Get "https://proxy.golang.org/github.c
om/pkg/errors/@v/v0.9.1.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/runtime/engine/config/oci/config.go:20:2: github.com/seccomp/containers-golang@v0.6.0: Get
"https://proxy.golang.org/github.com/seccomp/containers-golang/@v/v0.6.0.zip": dial tcp 172.217.160.113:443: i/o timeout../cmd/internal/cli/cgroups.go:21:2: github.com/shopspring/decimal@v1.3.1: Get "https://proxy.golang.org/g
ithub.com/shopspring/decimal/@v/v1.3.1.zip": dial tcp 172.217.160.113:443: i/o timeout../pkg/cmdline/cmd.go:16:2: github.com/spf13/cobra@v1.5.0: Get "https://proxy.golang.org/github.com/spf13/
cobra/@v/v1.5.0.zip": dial tcp 172.217.160.113:443: i/o timeout../pkg/cmdline/env.go:17:2: github.com/spf13/pflag@v1.0.5: Get "https://proxy.golang.org/github.com/spf13/
pflag/@v/v1.0.5.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/remote/endpoint/service.go:24:2: github.com/sylabs/json-resp@v0.8.1: Get "https://proxy.go
lang.org/github.com/sylabs/json-resp/@v/v0.8.1.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/app/apptainer/push.go:27:2: github.com/vbauerster/mpb/v7@v7.4.2: Get "https://proxy.golang.org
/github.com/vbauerster/mpb/v7/@v/v7.4.2.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/app/apptainer/push.go:28:2: github.com/vbauerster/mpb/v7@v7.4.2: Get "https://proxy.golang.org
/github.com/vbauerster/mpb/v7/@v/v7.4.2.zip": dial tcp 172.217.160.113:443: i/o timeout../pkg/util/capabilities/process_linux.go:15:2: golang.org/x/sys@v0.0.0-20220715151400-c0bba94af5f8: Get "
https://proxy.golang.org/golang.org/x/sys/@v/v0.0.0-20220715151400-c0bba94af5f8.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/util/interactive/interactive.go:23:2: golang.org/x/term@v0.0.0-20210927222741-03fcf44c2211
: Get "https://proxy.golang.org/golang.org/x/term/@v/v0.0.0-20210927222741-03fcf44c2211.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/remote/remote.go:27:2: gopkg.in/yaml.v2@v2.4.0: Get "https://proxy.golang.org/gopkg.in/yam
l.v2/@v/v2.4.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/util/shell/interpreter/interpreter.go:25:2: mvdan.cc/sh/v3@v3.5.1: Get "https://proxy.gola
ng.org/mvdan.cc/sh/v3/@v/v3.5.1.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/files/files.go:20:2: mvdan.cc/sh/v3@v3.5.1: Get "https://proxy.golang.org/mvdan.cc/s
h/v3/@v/v3.5.1.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/build/files/files.go:21:2: mvdan.cc/sh/v3@v3.5.1: Get "https://proxy.golang.org/mvdan.cc/s
h/v3/@v/v3.5.1.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/remote/credential/login_handler.go:20:2: oras.land/oras-go@v1.2.0: Get "https://proxy.gola
ng.org/oras.land/oras-go/@v/v1.2.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/remote/credential/login_handler.go:26:2: oras.land/oras-go@v1.2.0: Get "https://proxy.gola
ng.org/oras.land/oras-go/@v/v1.2.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/client/oras/oras.go:37:2: oras.land/oras-go@v1.2.0: Get "https://proxy.golang.org/oras.lan
d/oras-go/@v/v1.2.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/client/oras/oras.go:38:2: oras.land/oras-go@v1.2.0: Get "https://proxy.golang.org/oras.lan
d/oras-go/@v/v1.2.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/client/oras/oras.go:39:2: oras.land/oras-go@v1.2.0: Get "https://proxy.golang.org/oras.lan
d/oras-go/@v/v1.2.0.zip": dial tcp 172.217.160.113:443: i/o timeout../internal/pkg/client/oras/oras.go:40:2: oras.land/oras-go@v1.2.0: Get "https://proxy.golang.org/oras.lan
d/oras-go/@v/v1.2.0.zip": dial tcp 172.217.160.113:443: i/o timeoutmake: *** [apptainer] Error 1
看起来是编译过程中网络连接失败,造成部分库下载不到,导致编译失败。可以尝试如下方法解决问题:
如果您有一个本地存在的 RPM 包,您可以使用以下命令安装它:
sudo yum localinstall <filename>.rpm
non-setuid和setuid是关于Unix系统文件权限的术语。当一个程序被标记为
setuid时,当它被执行时,它的权限将从它的所有者变为它的执行者。这意味着即使普通用户执行了一个以setuid标记的程序,该程序仍将具有更高的权限,以便完成某些任务。相反,当一个程序被标记为
non-setuid时,该程序仅具有它的所有者的权限,即使它被普通用户执行。因此,在上面的代码中,
sudo yum install -y apptainer将安装一个没有setuid标记的版本,而sudo yum install -y apptainer-suid将安装一个以setuid标记的版本。通常,在安全方面,使用
non-setuid版本更安全,因为它仅允许该程序以其所有者的权限运行,而不允许它以执行者的权限运行。然而,如果程序需要更高的权限才能完成任务,则必须使用setuid版本。
sudo yum install -y apptainer这段代码是在执行在Red Hat系列的操作系统(如CentOS、Fedora等)上的软件包安装。yum是一个在Red Hat系列操作系统上进行软件包管理的工具。它允许安装、升级和删除软件包。
在这个代码中,
sudo命令用于在管理员权限下执行命令。yum install -y命令是安装软件包的命令,其中-y选项用于默认回答“是”,以便在没有用户交互的情况下安装软件包。最后,apptainer是要安装的软件包的名称。安装的软件包将被下载并存储在操作系统的软件仓库中,通常在
/var/cache/yum或/var/lib/yum目录下。安装后的文件将安装在系统的文件系统中,通常在/usr/目录下。
Install RPM from EPEL or Fedora
Multiple architectures of RPMs are available for Red Hat Enterprise Linux and Fedora.
First, on Red Hat Enterprise Linux derived systems enable the EPEL repositories like this:
$ sudo yum install -y epel-release
Then to install a non-setuid installation of Apptainer do:
$ sudo yum install -y apptainer
or for a setuid installation do:
$ sudo yum install -y apptainer-suid
Install from GitHub release RPMs
Alternatively, x86_64 RPMs are available on GitHub immediately after each Apptainer release and they can be installed directly from there. For the non-setuid installation:
$ sudo yum install -y https://github.com/apptainer/apptainer/releases/download/v1.1.5/apptainer-1.1.5-1.x86_64.rpm
For the setuid installation do above command first and then this one:
$ sudo yum install -y https://github.com/apptainer/apptainer/releases/download/v1.1.5/apptainer-suid-1.1.5-1.x86_64.rpmsingularity 安装与使用 – 简书 成功!!
echo ‘export PATH=~/.aspera/connect/bin/:$PATH’ >> ~/.bashrc
source ~/.bashrc singularity/INSTALL.md at master · apptainer/singularity · GitHub
如何直接使用sigularity 或者apptainer调用软件 virsorter2
Option 3
If you have apptainer (formerly known as singularity) installed (typical in HPC clusters), the following is the most convenient. Also use the option if you have issues with conda.
apptainer build virsorter2.sif docker://jiarong/virsorter:latestYou will get a file
virsorter2.sif, which is a singularity image that can be run like a binary executable file. You can use the absolute path of this file to replacevirsorterin commands for the rest of the tutorial. Also this image has the database and dependencies included, so you can skip the Download database and dependencies stephttps://github.com/jiarong/VirSorter2#download-database-and-dependencies below.
singularity使用
singularity build metawrap.sif docker://lmwbio/metawrap_1.3.2_binning_fastq_gz
INFO: Starting build...
Getting image source signaturessourmash 先安装pip 再用pip安装sourmashGitHub – sourmash-bio/sourmash: Quickly search, compare, and analyze genomic and metagenomic data sets.
Singularity 可以直接从 Docker Hub 下载镜像,您可以使用以下命令:
singularity pull docker://<DOCKER_IMAGE_NAME>:<TAG>
问题1. Singularity 可以直接从 Docker Hub 下载镜像速度太慢,我应该怎么办 问题2. 我可以先把docker镜像下载到本地,然后使用singularity来安装本地的镜像吗?具体应该怎么做
-
是的,您可以先下载 Docker 镜像到本地,然后使用 Singularity 安装本地的镜像。
具体操作如下:
docker save <IMAGE_NAME>:<TAG> -o <SAVE_FILE>.tar
docker save ubuntu:20.04 -o ubuntu_20.04.tar
singularity image.create <SINGULARITY_CONTAINER>.sif
例如:
singularity image.create ubuntu_20.04.sif
singularity import <SINGULARITY_CONTAINER>.sif <SAVE_FILE>.tar
singularity import ubuntu_20.04.sif ubuntu_20.04.tar
现在,您应该已经有了一个本地的 Singularity 容器,可以快速启动和运行。
windows系统安装dockerInstall on Windows | Docker Documentation
在 Windows 系统中,可以使用 Docker Desktop 软件下载 Docker 镜像。
首先,请确保已安装 Docker Desktop,并已启动 Docker 引擎。
然后,打开命令提示符或 PowerShell,输入以下命令:
docker pull lmwbio/metawrap_1.3.2_binning_fastq_gz
这将下载镜像到您的计算机上,并将其存储在 Docker 镜像库中。
稍后,您可以使用 docker run 命令启动该镜像。
例如:
docker run lmwbio/metawrap_1.3.2_binning_fastq_gz
metaphlan2
https://anaconda.org/bioconda/metaphlan2
metawrap
最终通过把metawrap下载到本地 然后使用conda安装本地软件
conda install --use-local *****container/Singularity.metawrap at master · hexmek/container · GitHub
在 CentOS 系统中安装 metaWRAP 使用 Singularity 可以通过以下步骤进行:
- 安装 Singularity:
Copy code
sudo yum install singularitybashCopy code
Bootstrap: docker From: ubuntu:16.04 %post apt-get update apt-get -y install wget wget https://github.com/bzhangHu/metaWRAP/releases/download/v1.0/metaWRAP-Linux.tar.gz tar -xvzf metaWRAP-Linux.tar.gz rm metaWRAP-Linux.tar.gz
- 创建 Singularity 容器:
pythonCopy code
sudo singularity build metawrap.sif metawrap.def
- 运行容器:
singularity run metawrap.sif这样,您就可以在 Singularity 容器中使用 metaWRAP 了。
megahit
SOAPdenovo2 soapaligner
GitHub – aquaskyline/SOAPdenovo2: Next generation sequencing reads de novo assembler.
vfdb
2021.3.17丨致病菌毒力因子(VFDB)数据库注释_穆易青的博客-CSDN博客_vfdb数据库
史上最详细的微生物扩增子数据库整理_刘永鑫Adam的博客-CSDN博客_greengene数据库
微生物多样性专题 | 扩增子测序分析实战(二)数据库整理之UNITE
原文地址:https://blog.csdn.net/qq_52813185/article/details/128916262
本文来自互联网用户投稿,该文观点仅代表作者本人,不代表本站立场。本站仅提供信息存储空间服务,不拥有所有权,不承担相关法律责任。
如若转载,请注明出处:http://www.7code.cn/show_14311.html
如若内容造成侵权/违法违规/事实不符,请联系代码007邮箱:suwngjj01@126.com进行投诉反馈,一经查实,立即删除!